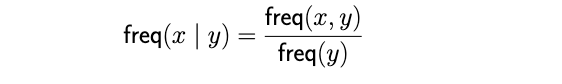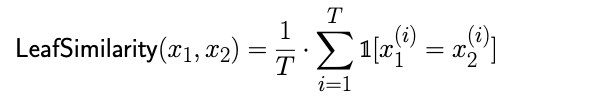- Binary code similarity (BCS) is an important part of training machine learning (ML) models to effectively analyze vast amounts of cybersecurity telemetry. However, BCS has historically focused on finding similarities among malicious examples rather than benign data, which limits its effectiveness.
- The CrowdStrike research team has released EMBERSim, a BCS dataset that builds on the existing EMBER dataset, with extended data tags and a new leaf similarity co-occurrence algorithm that accounts for both benign and malicious binaries.
- This innovative approach to qualifying similarity in cybersecurity improves the results of BCS in ML models, showing EMBERSim has the potential to improve malware detection and enable future work in this key research area.
CrowdStrike is constantly researching, working and innovating to stay at the cutting edge of threat detection and response. Recently, these efforts include EMBERSim, a large-scale dataset developed to address limitations in binary code similarity (BCS), improve malware detection and facilitate future work in this area.
In recent years, there has been a shift from heuristics to ML-based malware detection. Replicating both malicious and benign code is a common practice that usually results in similar examples, which are analyzed and curated to build new ML-based solutions or improve the performance of existing ML models. Being able to accurately identify similar examples is important in performing bulk analysis over the growing amounts of cybersecurity telemetry. BCS involves comparing two binary files (executables such as the .exe format) for similarities — a technique that is particularly important in cybersecurity because most malware is hidden within such files, making it more challenging to detect.
However, the CrowdStrike research team has identified a scarcity of data for similarity research in malware detection and observed a focus on quantifying similarity only in malicious examples and not in benign data. Given the vast majority of files processed will be benign, having the ability to label them as such would boost the ability to confidently perform a similarity search for an example file — a capability that becomes invaluable in the move toward automated analysis. We propose to address these limitations in BCS research. We are building our research on the existing EMBER dataset of Portable Executable (PE) files, which includes features and tags intended for malware classification.
Our contribution is threefold:
- We released EMBERSim, a dataset intended for BCS research that enhances the malware family (FAM) tags metadata (obtained via AVClass v1) in the original EMBER dataset with similarity information and malware class (CLASS) and behavior (BEH) tags as well as additional family (FAM) tags. This extended list of tags is determined via a co-occurrence algorithm that we have designed as part of our work.
- Based on the literature to date with applicability in the cybersecurity context, to our knowledge we are the first to repurpose an XGBoost malware classifier to quantify pairwise similarity at the leaf level.
- We are proposing a new extensive evaluation scheme to assess the effectiveness of the proposed leaf similarity technique — i.e., Top-K Selection and Relevance @ K. Using this method, we compared ourselves against an established off-the-shelf method for computing similarity in cybersecurity (i.e., ssdeep) and validated leaf similarity as a better alternative.
We documented this work in a paper that was peer-reviewed, accepted and presented in poster format at NeurIPS 2023. Through this blog post, we offer an overview of this work and invite readers to research the subject in more detail using our published paper, code and data as support.
Data Used for EMBERSim Project
We built our work on EMBER, a malware classification dataset of binary files in PE format. We used EMBER 2018, feature version 2, which consists of 1M total samples, out of which 800K are labeled (600K train and 200K test) and an extra 200K are unlabeled. The samples in train and test are perfectly split between “benign” and “malicious.” Additionally, the dataset follows a temporal split, wherein all test samples have a more recent timestamp compared to train samples. The EMBER dataset contains the SHA256s of the PEs, assigned ground truth and features.
In EMBERSim, aside from similarity information, we include extended tag information obtained via a co-occurrence-based tag enrichment procedure. We leverage AVClass v2 and obtain the following tag types:
- FAM (family) — e.g., grayware, backdoor, worm
- BEH (behavior) — e.g., XTRAT, zBot, Emotet
- CLASS (class) — e.g., infostealer, DDoS, inject
- FILE (file properties) — e.g., os:Windows, packed, packed:Themida
For each malicious sample in EMBER, we run AVClass to obtain their associated tags of the types enumerated above. We augment this initial list of tags using the co-occurrence procedure described in the upcoming section. A second set of steps is applied with the end goal of obtaining tag ranking, which we’ll use in the evaluation scheme to assess the performance of the binary code similarity method proposed. See Figure 1.
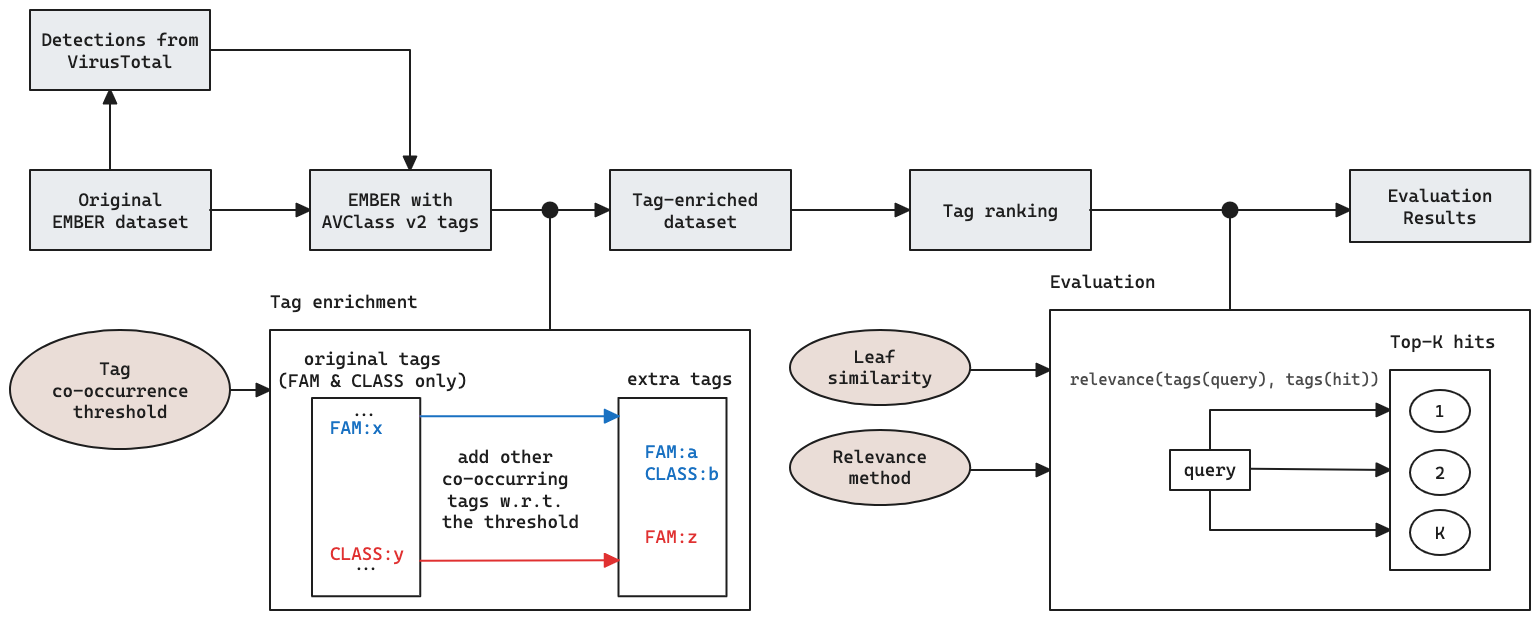
Figure 1. EMBERSim research pipeline, including the tag enrichment based on co-occurrence, ranking and how the tags are leveraged for evaluating the leaf similarity method proposed as part of this work
Metadata Augmentation
Tag Enrichment via Co-Occurrence
For each sample in EMBER, we use its SHA256 to query VirusTotal (VT) and then run AVClass v2 to obtain tags with their corresponding confidence scores across multiple vendors. AVClass provides co-occurrence statistics for tag pairs (out of which we can build a co-occurrence matrix). We leverage this information to enrich the set of tags of the malicious samples in EMBER by adding the co-occurring ones above a frequency threshold. Given that we target similarity research, the main goal with this enhancement is being able to find samples with shared characteristics, even when it comes to different families. Algorithm 1 displays the step-by-step tag enrichment procedure followed.

Algorithm 1. Tag enrichment procedure based on leveraging the AVClass sourced co-occurrence statistics for the examples in EMBER
Tag Ranking for Evaluation
Following the procedure described in Algorithm 2, we obtain a tag ranking for each sample in EMBERSim. The end goal is to use the tag ranks in performing the Relevance @ K evaluation (i.e., how relevant to the query samples are the top-K most similar retrieved items?).
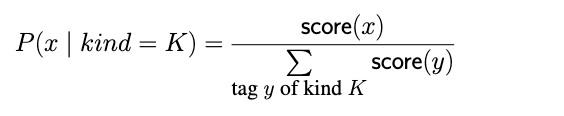
In Algorithm 2, P represents a normalized measure of confidence (by kind), which is derived from the AVClass tag scores and defined as below:
Additionally, the freq(x, y) variable used in computing the rank score represents the common co-occurrence of frequency of tags x and y, from which we can derive a relative occurrence frequency of tag x given tag y.
Leaf Prediction Similarity Method
Given a trained tree ensemble with a total of T trees and two samples x1 and x2, we define the similarity of x1 and x2 as the leaf similarity between them in the context of the aforementioned tree ensemble.
Although the method can be applied to any type of tree ensemble, in our experiments we opt for eXtreme Gradient Boosting (XGBoost). Therefore, we train an XGBoost-based malware classifier over the original features released alongside the EMBER dataset. The XGBoost malware detector trained has similar capabilities with the LightGBM baseline described in the original EMBER paper. Our model uses the following hyperparameter set: max_depth = 17; η = 0.15; n_estimators = 2048; colsample_bytree = 1. The performance of the malware classifier itself is subject to hyperparameter optimization and of lesser importance for our research.
With a competitive XGBoost-based malware detector, we continue by extracting leaf predictions for the two samples, which are the subject of our similarity assessment. The similarity score represents the fraction of the trees in which both samples fall into the same leaf node. The intuition behind this idea is that if two samples consistently end up in the same leaf node across multiple decision trees, it suggests that they have similar characteristics or share common patterns in their features.
Example
Figure 2 is an example considering three samples (sample 1, sample 2 and sample 3) and an ensemble of three trees (Tree #1, Tree #2 and Tree #3), with the respective paths through the trees highlighted.
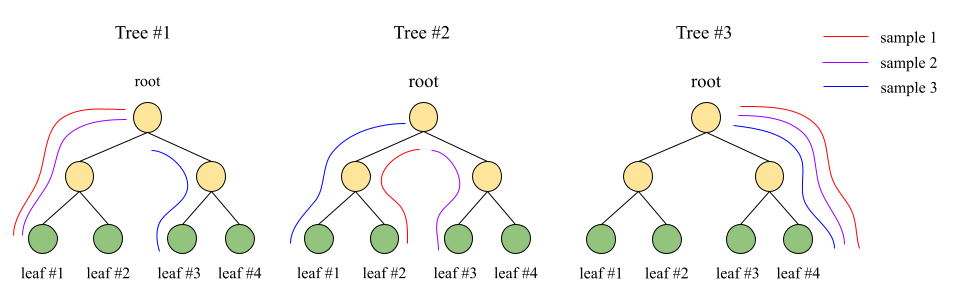
Figure 2. Example of how leaf predictions are considered in each of the three trees of an ensemble model for three examples that we need to assess similarity-wise via the leaf similarity technique
From the paths drawn in Figure 2, we can infer that the leaf predictions for these samples are [1, 2, 4], [1, 3, 4] and [3, 1, 4] for samples 1, 2 and 3, respectively. In this case, the LeafSimilarity score for sample 1 and sample 2, given the formula above, is 0.66.
Evaluation
We employ two different evaluation scenarios, with the end goal of validating the novel similarity method based on leaf predictions. As part of the evaluation procedure, we leverage the tag augmentations and ranking obtained using the algorithms described in the Metadata Augmentation section above. Figure 3 displays the two evaluation paths that were considered — namely, the Top-K selection and Relevance @ K evaluation. In the following subsections, we describe the two methods in more detail.
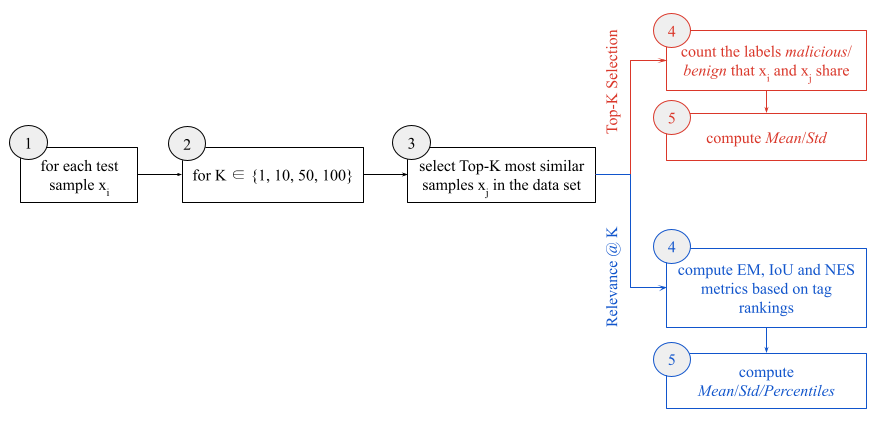
Figure 3. Evaluation workflow with two different ramifications for the two scenarios considered, namely Top-K selection and Relevance @ K evaluation
Top-K Selection
We conduct a homogeneity-based comparative evaluation of the leaf similarity method proposed in our paper and ssdeep, which is well established and extensively used in the cybersecurity domain. The Top-K selection evaluation is based on counting the (malicious/benign) ground truth labels that a query and its most similar K (EMBER) samples share. We report the mean and standard deviation, as shown in Table 1.
Label Homogeneity for ssdeep | Label Homogeneity for Leaf Similarity | |||||||||||
| K | Benign | Malicious | All | Benign | Malicious | All | ||||||
| Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | |
| 1 | 0.51 | 0.49 | 0.80 | 0.39 | 0.66 | 0.47 | 1 | 0 | 1 | 0 | 1 | 0 |
| 10 | 3.55 | 4.34 | 7.31 | 4.16 | 5.43 | 4.65 | 9.68 | 1.13 | 9.80 | 0.97 | 9.74 | 1.06 |
| 50 | 12.31 | 19.24 | 32.59 | 22.12 | 22.45 | 23.07 | 47.10 | 7.34 | 48.40 | 6.17 | 47.75 | 6.81 |
| 100 | 19.84 | 34.82 | 61.00 | 45.22 | 40.42 | 45.31 | 92.80 | 16.01 | 96.28 | 13.22 | 94.53 | 14.79 |
Table 1. Comparative evaluation of leaf similarity and ssdeep, considering the Top-K selection scenario, where K is in {1, 10, 50, 100}. We report the mean and standard deviation obtained from considering the benign, malicious and all tags. Mean values closer to K and Std values closer to 0 indicate better performance.
Our analysis shows a poor overall performance for the (rather restrictive) ssdeep method, with marginally better scores for the malicious samples than for the clean ones. The results can indicate an arbitrary level of success in identifying the two. Leaf similarity outperformed ssdeep, achieving better results for both malicious and benign queries, showcasing its effectiveness in accurately identifying and distinguishing between the two.
Relevance @ K Evaluation
Aside from the Top-K selection evaluation, we conducted a second type of evaluation that involves assessing the relevance of the retrieved results in the tags enrichment scenario. To perform the Relevance @ K Evaluation, we leverage the tag ranking, which serves as a reference for determining the relevance of the retrieved samples. We then check whether the tag ranking of a query sample is consistent with the Top-K most similar samples retrieved by our model using various scoring mechanisms, which we enumerate and briefly describe below:
- EM (Exact Match) — outputs 1 if the inputs are equal. This is the strictest comparison method, serving as a lower bound for that performance.
- IoU (Intersection over Union) or Jaccard index — disregards element ordering and focuses solely on the presence of the items in the inputs.
- NES (Normalized Edit Similarity) — this function, based on Damerau-Levenshtein distance (DL) for strings, allows us to penalize differences in rank.
The use of these mechanisms allowed us to measure the relevance and accuracy of our approach.
To construct the query and knowledge base subsets, we consider two scenarios: unsupervised labeling and counterfactual analysis. For the counterfactual analysis, we leverage the train-test temporal split (i.e., timestamp(test) > timestamp(train)) in EMBER to simulate a real-world production-like scenario, where we use unseen but labeled data (i.e., the test set) to query the knowledge base. With the Unsupervised labeling scenario, given the unlabelled EMBER data, we aim to identify similar samples within the knowledge base, with the purpose of assigning a provisional label or tag.
In the Unsupervised labeling scenario, we achieved excellent results. Our model demonstrated the ability to accurately match queries with hits based on their tags, further validating the effectiveness of our approach. Similarly, In the counterfactual analysis scenario, we observe that leaf similarity achieves very good results in terms of matching queries and hits according to their tags. This indicates that our approach can successfully identify and retrieve relevant samples based on the leaf predictions.
Evaluation Results for Mean Average Precision | ||||
| Top K | Unsupervised | Counterfactual | ||
| Benign | Malicious | Benign | Malicious | |
| 1 | 0.986 | 0.716 | 0.996 | 0.960 |
| 10 | 0.985 | 0.753 | 0.985 | 0.895 |
| 50 | 0.973 | 0.714 | 0.967 | 0.812 |
| 100 | 0.966 | 0.698 | 0.957 | 0.783 |
Table 2. Aggregated results for the Relevance @ K evaluation in both the unsupervised and counterfactual testing scenarios
One Team, One Fight
CrowdStrike researchers, threat analysts and data scientists work tirelessly to stay ahead of adversaries. This focus on research, innovation and thought leadership includes numerous projects — such as EMBERSim — and extends to industry leadership through involvement in organizations like the MITRE Engenuity Center for Threat-Informed Defense, where CrowdStrike is a Research Partner. The results of our research efforts and the information that we share lead to improving defenses globally against the latest and most sophisticated adversary tactics and malware. For our customers, these efforts mean that the AI-native CrowdStrike Falcon® platform always delivers best-in-class cybersecurity protection.
Additional Resources
- Learn more about the Falcon platform by visiting the product webpage.
- Learn more about CrowdStrike endpoint detection and response by visiting the CrowdStrike Falcon® Insight XDR webpage.
- Test CrowdStrike next-gen antivirus for yourself. Start your free trial of CrowdStrike® Falcon Prevent™ today.




![Helping Non-Security Stakeholders Understand ATT&CK in 10 Minutes or Less [VIDEO]](https://assets.crowdstrike.com/is/image/crowdstrikeinc/video-ATTCK2-1)
![Qatar’s Commercial Bank Chooses CrowdStrike Falcon®: A Partnership Based on Trust [VIDEO]](https://assets.crowdstrike.com/is/image/crowdstrikeinc/Edward-Gonam-Qatar-Blog2-1)
![Endpoint Protection and Threat Intelligence: The Way Forward [VIDEO]](https://assets.crowdstrike.com/is/image/crowdstrikeinc/GK-Blog_Images-1)